Statitical Method Used to Describe Bacteria Abundance
The FCM count ranged from 391 10 8 to 569 10 8 cells g 1 soil for crop field and was 669 10 8 cells g 1 soil for grassland. And the bacterial top 15 and fungal top 10 most abundant classes with environmental altitude mean annual air temperature MAT mean annual soil temperature.
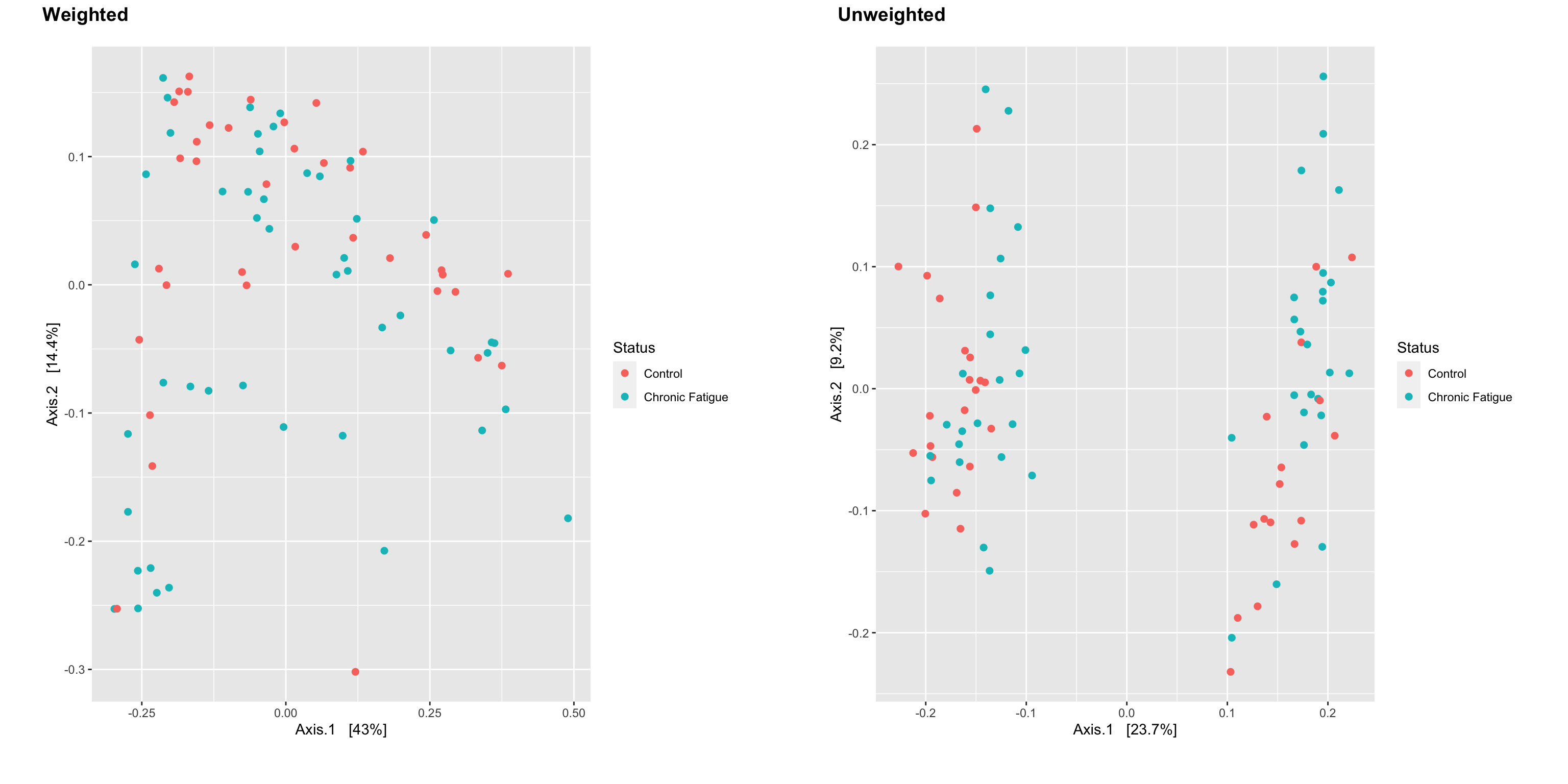
Introduction To The Statistical Analysis Of Microbiome Data In R Academic
The top layer obviously favors oxygen-loving microbes whereas the bottom layer which is submerged in water selects for oxygen sensitive strains.
. Statistical Analysis of Microbiome Data in R by Xia Sun and Chen 2018 is an excellent textbook in this area. Researchers may further wish to determine through statistical testing which specific bacteria are significantly differentially abundant between two ecosystems. Although there are various methods used to perform data analysis given below are the 5 most used and popular methods of statistical analysis.
Veined rough dull wrinkled or shriveled glistening. Various terms used in this paper are summarized in Table 1. In contrast fewer methods are available for estimating abundance when the probability of presence p p is less than 1 which is the focus of this work.
Metastats p and q values students t-test. Here we aimed to optimize the use of flow cytometry FCM to assay bacterial. Gammaproteobacteria had the highest abundance 1528 followed by unidentified_Gemmatimonadetes 890 and Thermoleophilia.
STATISTICAL METHODS 1 STATISTICAL METHODS Arnaud Delorme Swartz Center for Computational Neuroscience INC University of San Diego California CA92093-0961 La Jolla USA. The different diversity characteristics of bacterial and fungal communities. Organic integrated with TRC or without tillage RTRC and conventional.
Fecal coliform bacteria inhabit the intestinal tracks of animals. Alternatively additional methods such as double-observer methods could be used to estimate group size when p d 1. Absolute abundances tend to be greatly right-skewed in distribution so log-transformation will be useful if statistical methods that assume normal distribution are used.
Also include descriptive terms for any other relevant optical characteristics such as. 10 You have conducted a spatial study in the ocean using metagenomics and found that there. The notations described in statistical methods are summarized in.
Statistics represents that body of methods by which. Multiple-correlation analyses using Pearsons method were used to relate archaeal bacterial and fungal abundances. There are a variety of ways to enumerate the number of bacteria in a sample.
A Name a method you would use. The direct count method for enumerating bacteria in natural environments is widely used. Ri and Rc are the relative.
Bacterial colonies are frequently shiny and smooth in appearance. After calculations are used to detirmine the sizeUsually the procedure is done indirectly with a series of dilutions making it possible to estimate the number of bacteria in the original sample. Use predictive analysis to predict future trends and events likely to happen.
Up to 24 cash back Most methods of counting are based on indirect or direct counts of tiny samples. We used this FCM method to compare bacterial abundance measured in four types of crop management systems. The plate count method or spread plate relies on bacteria growing a colony on a nutrient medium.
Instead they should have referred to it as observed abundance. The relative abundance of each COG is presented together with significance values likelihood that difference in abundance is due to chance alonecomputed using several statistical procedure. 1 Z Ii T Ii Ri Z T Rc T 1 Ri Ri Rc Ii where Ii is the absolute abundance of ISS HAAQ-GFP in each inoculated treatment.
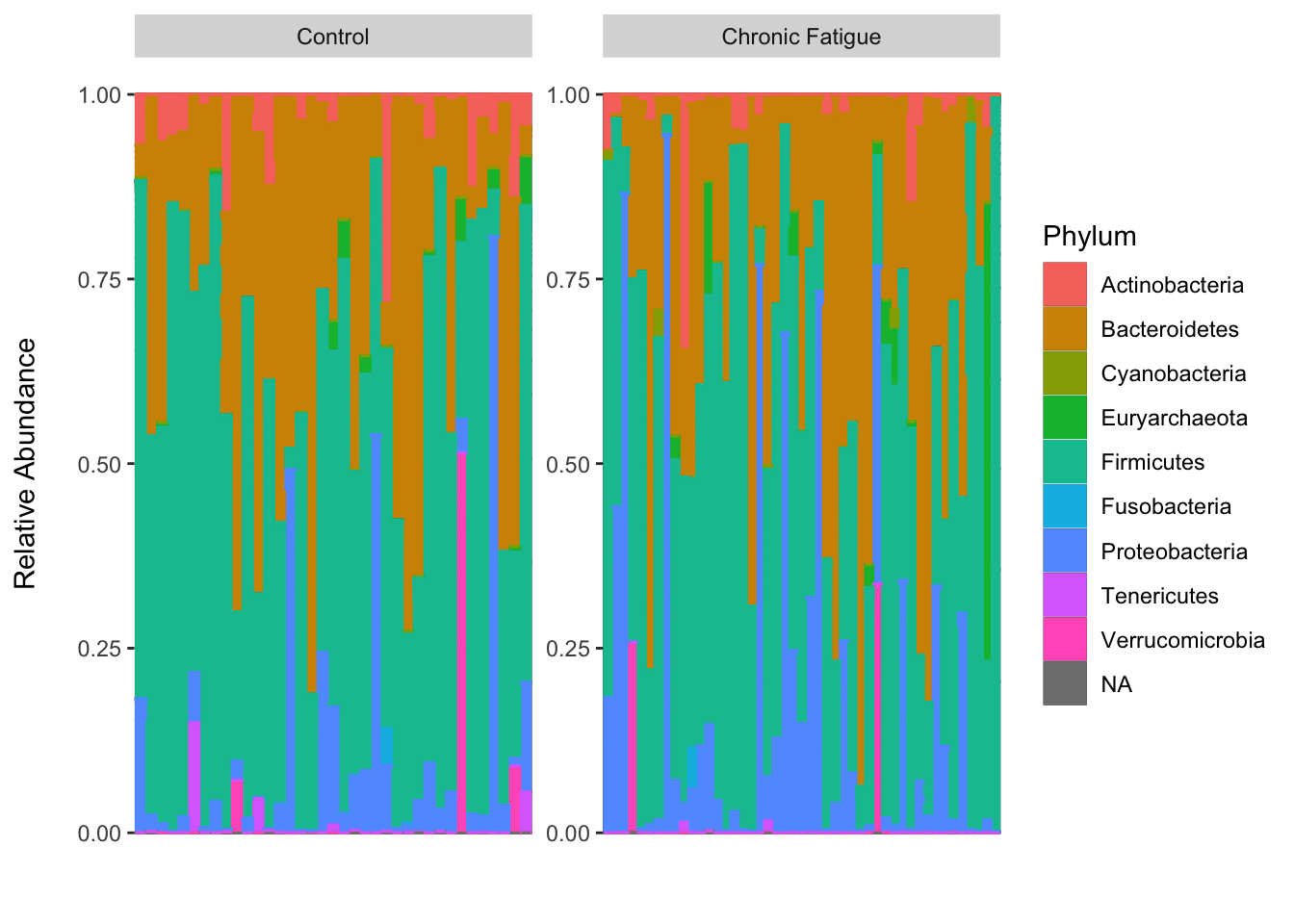
Figure 2b shows the top ten bacterial classes in the rhizosphere soil. For those looking for an end-to-end workflow for amplicon data in R I highly recommend Ben Callahans F1000 Research paper Bioconductor Workflow for. By growing hundreds of bacterial communities collected from a natural aquatic environment rainwater-filled tree holes under the same environmental conditions we show that negative statistical interactions among abundant phylotypes drive variation in broad functional measures respiration metabolic potential cell yield whereas positive interactions between rare.
With the relative abundance of the Escherichia genus and the absolute abundance of ISS HAAQ-GFP the absolute abundance of the total soil indigenous bacteria was calculated using the following equation. Other methods such as viable plate counts can also be used for determining bacterial growth. While the indicator bacteria are typically not pathogens they indicate that the water has become.
Mean or average mean is one of the most popular methods of statistical analysis. Relative abundance tells us how many percentages of the microbiome is made up of a specific organism eg. Color It is important to describe the color or pigment of the colony.
Statistical methods inference models clinical software bootstrap resampling PCA ICA Abstract. The sequencing solutions we use to profile the microbiome of a sample provide information on the relative abundance of the microorganisms in each sample. Despite its importance methodological constraints hamper our ability to assess bacterial abundance in terrestrial environments.
The abundance of fecal coliform bacteria are used as an indicator of fecal contamination of both drinking water and recreational water ie swimming shellfishing. People am also facing the same challenge on which statistical method to use I am interested in species description and distribution over four agro-ecological zones at different stages of crop. For the statistical analysis of bacterial abundances relative or absolute the distribution of the data should be considered.
This is the most common method used to rapidly estimate bacterial numbers. B Briefly describe 1 problem that may cause your results to be biased. This is the most frequently used method.
Coli make up 1 or 10 of the total amount of bacteria detected in a sample. This process is known as differential abundance testing. A viable cell count allows one to identify the number of actively growingdividing cells in a sample.
There are many great resources for conducting microbiome data analysis in R. Bacterial abundance is a fundamental metric for understanding the population dynamics of soil bacteria and their role in biogeochemical cycles. A Name a method you would use to do this.
Measuring the turbidity of the culture solution can be used in estimating numbers of bacterial cells if a growth curve for the conditions used has already been established. This paper analyzes the sources of. An alternative method used with the serial dilutions is called pour plating where the crust-water solution can be poured to differentiate among microorganisms based on their oxygen dependence.
9 You are investigating the relative abundance of Bacteria and Archaea in a marine cold seep. Other surface descriptions might be.

Bacterial Abundance Types Bacterial Abundance Types Include Symmetric Download Scientific Diagram

Introduction To The Statistical Analysis Of Microbiome Data In R Academic
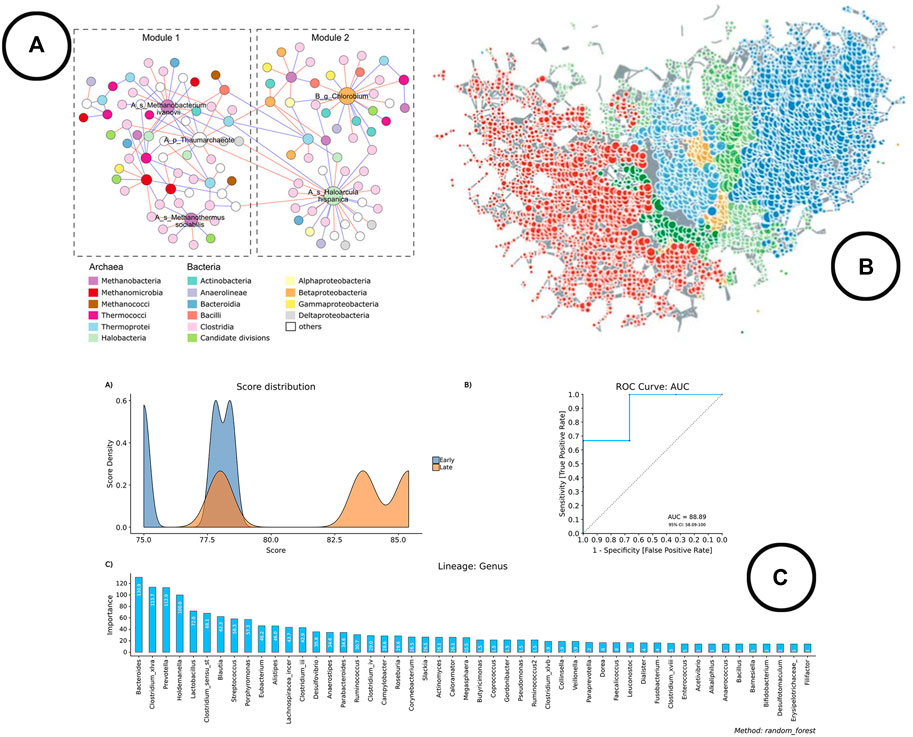
Frontiers Exploring The Microbiome Analysis And Visualization Landscape Bioinformatics
Comments
Post a Comment